Dr Kovanich utilizes mass-spectormetry based proteomics approches to map and visualize protein interaction networks.
Tel: 66 (0) 2441-9003 – 7 Ext. –
Email: duangnapa.kov mahidol.ac.th
mahidol.ac.th
Ph.D. (Biomolecular Mass Spectrometry and Proteomics), Utrecht University, 2013
Academic Program(s)
Molecular and Integrative Biosciences
Systems Biosciences
Affinity purification coupled with mass spectrometry (AP–MS) and proximity-dependent biotinylation identification (BioID) methods have made substantial contributions to interaction proteomics studies. We have developed a GFP-tag AP-MS workflow to study the virus-host interactions of several medically and economically important viruses in Thailand. We are currently developing a proteomics platform for the dynamic visualization of signalosome landscape, where we combine the CRISPR-Cas9 genome editing with the BioID system. Cells will be engineered to produce the biotin ligase-tagged signaling protein of interest and BioID will be performed following the activation of the signaling pathway in a time-course manner. When paired with quantitative proteomics approach, the assay will be able to provide snapshots of both core and transient protein landscape of the signaling pathway over time.
1.Expanding the Flaviviral NS5 Interactome: A Proteomic Insight into Host-Pathogen Interactions Kovanich D, Saisawang C, Sittipaisankul P, Ramphan S, Kalpongnukul N, Somparn P, Pisitkun T, Smith DR. Analysis of the Zika and Japanese Encephalitis Virus NS5 Interactomes. J Proteome Res. 2019 Aug 2;18(8):3203-3218.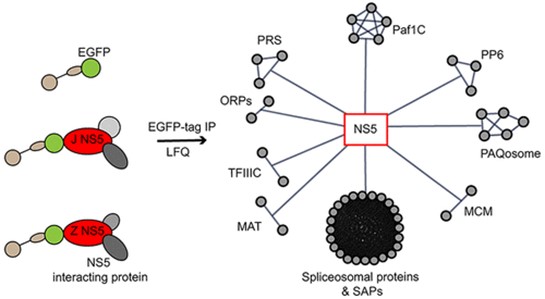
Mosquito-borne flaviviruses, including dengue virus (DENV), Japanese encephalitis virus (JEV), and Zika virus (ZIKV), remain significant global health threats. Among flaviviral proteins, nonstructural protein 5 (NS5) is the largest, most conserved, and enzymatically vital component of the viral replication complex, making it an attractive target for broad-spectrum antiviral strategies. Understanding NS5-host protein interactions is critical for unveiling the molecular mechanisms that underpin flavivirus infection and replication.
In this study, our team conducted a comprehensive proteomic analysis to map the JEV- and ZIKV-NS5 interactomes. Using EGFP immunoprecipitation and label-free quantitative mass spectrometry, we identified 137 NS5 interactors, with a pronounced enrichment in spliceosomal and transcriptional regulatory proteins, suggesting that NS5 plays a key role in modulating host RNA processing. Notably, we identified the transcription complex Paf1C and phosphatase 6 as conserved NS5-associated complexes, providing novel mechanistic insights into viral replication. Interestingly, PAF1, a core component of Paf1C, exhibited opposing roles in JEV and ZIKV infections, emphasizing virus-specific host dependencies. Our findings contributed to subsequent discoveries by other research groups, who later characterized the functional roles of Paf1C in flavivirus infection. This independent validation underscores the broader significance of our proteomic dataset in unraveling critical host pathways manipulated by flaviviruses.
https://www.mdpi.com/1999-4915/15/5/1032
https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1010100
4. Kovanich D, Ketsuwan K, Hengphasatporn K, Thepparit C, Sittipaisankul P, Wongkongkathep P, Sirisereewan C, Techakriengkrai N, Nedumpun T, Shigeta Y, Pisitkun T, Suradhat S. Analysis of the Porcine Reproductive and Respiratory Syndrome Virus Nucleocapsid Interactome. J Proteome Res. 2025 Nov 7;24(11):5390-5411.
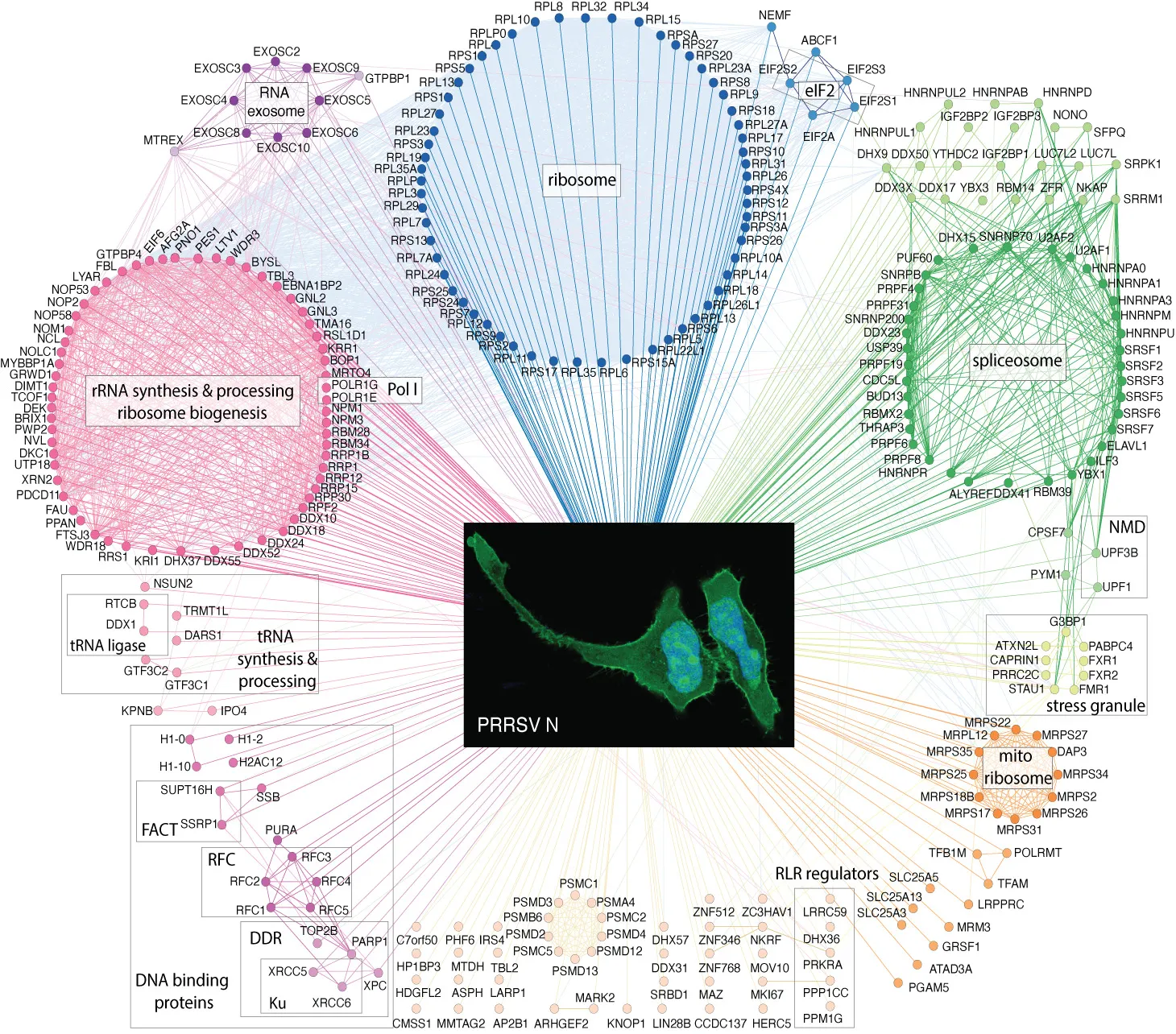
Porcine reproductive and respiratory syndrome virus (PRRSV) remains a major threat to the swine industry worldwide. Our lab applied advanced proteomics to explore host proteins interacting with the viral nucleocapsid (N) protein, revealing how this key viral component engages cellular machinery. Notably, we discovered a previously unrecognized pool of N protein on the cell surface, suggesting new functional roles beyond its canonical nuclear and cytoplasmic localization. Building on this, we are now characterizing the host cytosolic proteins that associate with N at the cell surface, aiming to uncover novel mechanisms of viral manipulation and potential intervention points. This work highlights the richness and complexity of virus-host interactions and illustrates our lab’s commitment to revealing hidden layers of cellular networks through integrative proteomic approaches.
![]()




